Homework 10
Up to the Phylogenetics main page
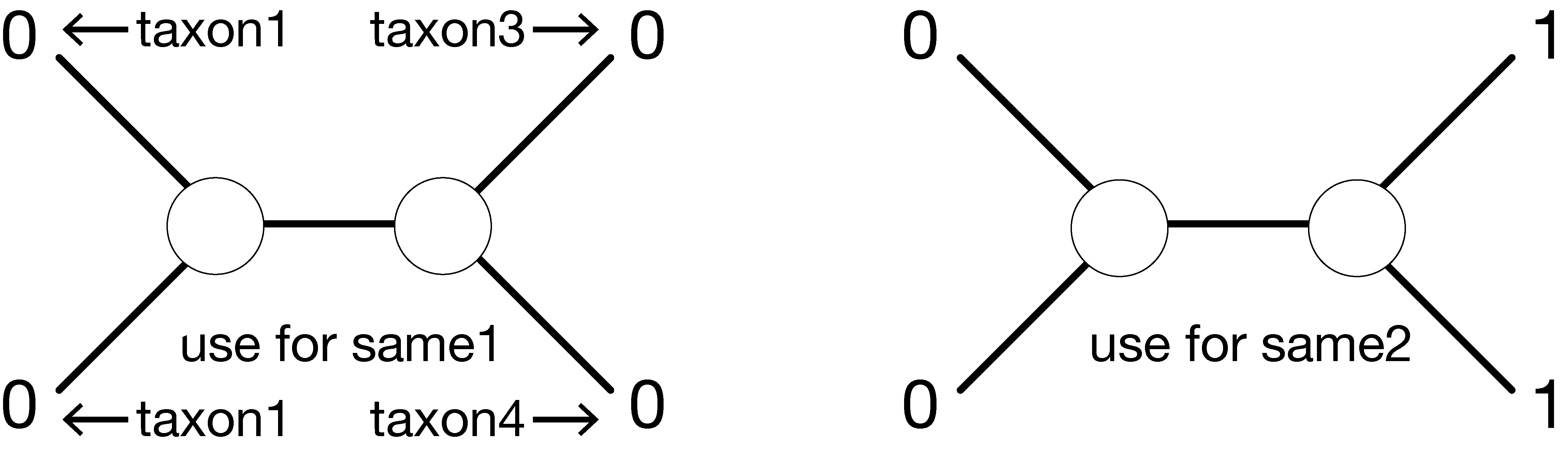
Conditioning on variability in the Mk model
The python script below will be modified by you to compute the site likelihoods of a 4-taxon tree for 2 characters using the Mk 2-state model. The first of the two characters is constant and the second character is variable. You will use the site likelihood for the constant character to condition the site likelihood for the variable character on the fact that it is variable. You will check your results using PAUP*.
Be sure to ask for help if you get stuck.
What to do
Login to your student account on the cluster and create a file named hw10.py using nano containing the code below. Edit the hw10.py script in nano, replacing the xxxx placeholder with the correct formula. Assume the data for characters 1 and 2 are as shown in Figure 1.
from math import exp, log
# You do *not* need to modify this function
# same_list holds the number of edges with the same state at both ends
# over all 4 possible combinations of ancestral states
# v is the edge length (same for all 5 edges in the tree)
def sitelike(same_list, v):
probsame = 0.5 + 0.5*exp(-2.*v)
probdiff = 0.5 - 0.5*exp(-2.*v)
like = 0.0
for n in same_list:
nsame=float(n)
ndiff=float(5-n)
like += 0.5*(probsame**nsame)*(probdiff**ndiff)
return like
# 00 01 10 11 <-- ancestral state combinations possible
same1 = [ 5, 2, 2, 1] # no. edges with same state at both ends
same2 = [ 3, 4, 0, 3] # in tree 1 (same1) and tree 2 (same2)
# Compute the site likelihood for each of the two characters
# by calling the function sitelike (we'll assume v = 0.1 for
# all edges.
v = 0.1
like1 = sitelike(same1, v)
like2 = sitelike(same2, v)
print('Mk model (edge lengths all %.1f):' % v)
print(' likelihood (char. 1) = %.5f' % like1)
print(' likelihood (char. 2) = %.5f' % like2)
# TODO: calculate likelihood for character 2 conditioned on variability
like2cv = xxxx
print('Mk model (edge lengths all %.1f, conditioning on variability):' % v)
print(' likelihood (char. 2) = %.5f' % like2cv)
print(' log-likelihood (char. 2) = %.5f' % log(like2cv))
Ensure that you have access to python3 on the cluster by loading this module:
module load python/3.8.1
Run the hw10.py script as follows:
python3 hw10.py
The sitelike function
Near the top of your hw10.py file is the definition of a function named sitelike. The function sitelike takes 2 arguments. The first,same_list, is a list of 4 numbers representing the number
of edges (of the 5 total) in which the state is the same
at both ends. There are 4 such values because there are 4
possible ancestral state combinations (00, 01, 10, 11) for 2 states (0 and 1).
The second is v, which is the edge length to be used for all 5
edges.
Checking your answer
Check your answer using PAUP*. You can get the data for this homework into PAUP* by creating a nexus file named hw10.nex. Your data file will be very similar to the one used for homework assignment 6 (rate heterogeneity) with the exception that the datatype specified in the format statement should be set to standard (not dna) and the states in the data matrix will be 0s and 1s (not As, Cs, Gs, and Ts). Dredge up your copy of your hw6.nex file and simply change the data and paup block commands.
The paup block you should use is provided below:
begin paup;
set crit=like;
[!
***************************************
*** Not conditioning on variability ***
***************************************
]
lset nst=1 genfreq=equal condvar=no;
lscores 1 / sitelike userbrlen;
[!
***********************************
*** Conditioning on variability ***
***********************************
]
exclude 1;
lset nst=1 genfreq=equal condvar=yes;
lscores 1 / sitelike userbrlen;
quit;
end;
The paup block above has two sections. The first section computes the likelihood for each character without conditioning on variability. (Note that PAUP* requires you to use genfreq rather than basefreq if the datatype is standard.) The exclude 1 statement in the second part is needed in order to exclude the first character (because constant characters are not allowed if condvar is set to yes).
It turns out that the most recent version of PAUP* installed on the cluster (4a166) will not work for this homework assignment. You can download version 168 to your account from the PhyloSolutions web site using curl:
curl -O https://phylosolutions.com/paup-test/paup4a168_centos64.gz
The file you download will have a gz filename extension, which indicates that it has been compressed using the program gzip. To uncompress it, use gunzip:
gunzip paup4a168_centos64.gz
This will simply replace the file named paup4a168_centos64.gz with a file named paup4a168_centos64. Finally, you will need to assure the operating system that this file is indeed executable by changing the mode of the file using the chmod command (the +x is code for “make it executable”):
chmod +x paup4a168_centos64
(Linux makes you go through this last step so that it will be entirely your fault if you execute a file you downloaded from the internet and it turns out to be a malicious computer virus.) Now you should be able to start PAUP/* like this:
./paup4a168_centos64
(You can also download version 4a168 of PAUP* to your laptop from the PhyloSolutions web site and run it on your laptop if that is more convenient.)
What if my answer differs from PAUP*’s answer?
First, make sure you are getting the correct site likelihoods without conditioning on variability.
Second, if you are getting the right site likelihoods when not conditioning on variability but not matching when you do condition on variability, be sure you are correctly calculating the probability of the site being variable. This value is 1 minus the probability that a site is constant, and remember that there are two ways that a site can be constant (either all 0s or all 1s at the leaves). You have calculated the probability that a site has all 0s (this is stored in the like1 variable), and the probability of a site having 1 at every leaf is the same, so all you need to do is multiply the value stored in like1 by 2 to get the probability that a site is constant.
What do I turn in?
Please send me a direct message in Slack and attach both your python program hw10.py as well as your hw10.nex file.